Advanced Genetic Algorithms
The
program AGA may be used to solve the great majority of
the one-criterion optimisation problems. Short description of this program is
presented below on the basis of two variables function optimisation.
(1) ![]()
given by
(2) ![]()
The
smallest values
![]()
The easiest
way to find that minimum is to write a script file in the following format
[VARIABLES]
x = real [0; 3.1415]
y = real [0; 3.1415]
[FUNCTION]
analytical = 1
[EQUATIONS]
f = -5.*sin(x)*sin(y) - sin(5*x)*sin(5*y)
[SHOW]
arrange = 1
[OPTIONS]
maximisation = 0
loop_counter = 100
max_iteration = 100
[SELECTION]
tournament = 1
tournament_size = 3
[CROSSOVER]
probability = 0.7
arithmetical = 1
[MUTATION]
probability = 0.15
nonuniform = 1
[POPULATION]
size = 30
constatnt = 1
[PLOTS]
Convergence = Average;CurrentMin
D = Discrepancy
Different_vs_2^Entropy =
Different;2^Entropy
Save it as ‘test.txt’ and you can run it writing in
your command line ‘aga test.txt’. Then you shall see something like this
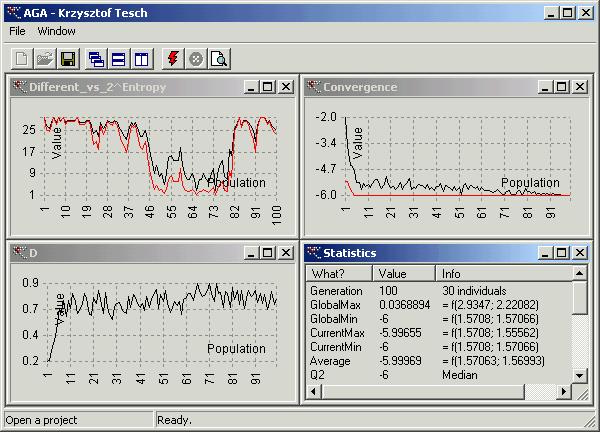
AGA found the infimum
called ‘GlobalMin’ (see window named ‘Statistics’) ![]() . It is very close to the exact value. If you do not want to
use the command line then fire up AGA and you see window
. It is very close to the exact value. If you do not want to
use the command line then fire up AGA and you see window
Choose option ‘Open’ from menu ‘File’ and then find
your file ‘test.txt’ and choose ‘Open’.
Instead of writing script file you can define your
optimisation problem manually. To do this choose menu ‘File’
-
‘New...’
– click here if you want to define your problem
-
‘Open...’
– open your problem definition from file. There are two types of file
-
*.txt
– script files (text)
-
*.aga
- AGA internal files (binary)
-
‘Close
all’ – closes all the windows and finish your optimisation
-
‘Save’
– save current problem
-
‘Save
as…’ – save current problem with a different name
-
‘Run’
– run your optimisation process
-
‘Terminate…’
– terminate your running process
-
‘About…’
– short note about the author
-
‘Exit’
– exit application
when you choose option ‘New’ from menu ‘File’ you will
se dialog box called ‘Initialise’. Dialog box ‘Initialise’ is composed of five
pages. First page ‘Variables’ is used to declare optimisation variables (in the
case of function optimisation – independent variables).
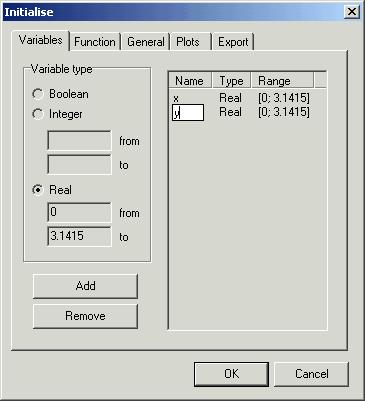
There are
three types of variables:
-
‘Boolean’
– logical variable. Two values are possible: 0 (false) and 1 (true).
-
‘Integer’
– integer variable. Values from the subset of integer numbers ![]() are possible. If an
integer variable has been chosen it is necessary to specify the range of the
desired subset. There are two edit windows: ‘from’ and ‘to’.
are possible. If an
integer variable has been chosen it is necessary to specify the range of the
desired subset. There are two edit windows: ‘from’ and ‘to’.
-
‘Real’
– floating point variable. Values from the subset of floating point numbers ![]() are possible. In this
case also it is necessary to specify the range of the desired subset. There are
two edit windows: ‘from’ and ‘to’.
After filling them the range of the subset is given by
are possible. In this
case also it is necessary to specify the range of the desired subset. There are
two edit windows: ‘from’ and ‘to’.
After filling them the range of the subset is given by ![]() .
.
When the
edit windows ‘from’ and ‘to’ are filled, the next step is to add variables to
the list. To do this: use a button ‘Add’. If you want to remove a variable from
the list then highlight it and push ‘Remove’ button. If you want to name your
variable just click on the empty place in the column ‘Name’. If the name is not
specified the program will assume default name (x1, x2, …). In our case we can
name our variables: x and y.
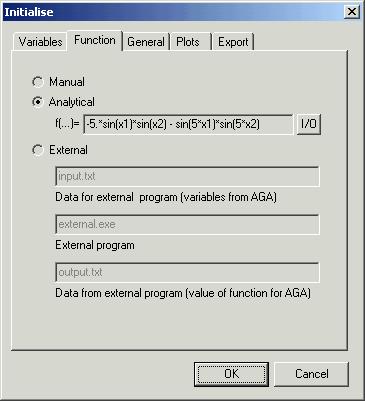
Second
page control ‘Function’ of the dialog box ‘Initialise’ determines the entry
method for the value fitness function. There are three possibilities:
-
‘Manual’
– enter individual values by hand
-
‘Analytical’
– you can specify analytical equation
-
‘External’
– automatically by external program (*.exe or *.bat file).
Manual
entry of the value fitness function is required for all the values that are not
calculated yet during the optimisation process step.
We
decided to deliver value fitness function by analytical equation. To do this
input in edit box ‘f(…)=’:
-5.*sin(x)*sin(y) - sin(5*x)*sin(5*y)
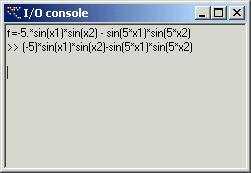
If
description of the function is more complicated the you have to push button
‘I/O’ – that will open I/O console. For more precision description see ‘I/O
Console’ paragraph. The function must be called ‘f’.
If you decide to deliver value fitness function
from an external program, you must specify the name of the text file into which
AGA
will send independent variables that are generated by GA. The external program
must calculate value of the function for these variables (‘Data for external
program (variables from AGA)’). Default name of this file is ‘input.txt’. The
name of executable (*.exe) program or *.bat file which is going to calculate
these values must also be specified (‘External program’). The external program
has to save the calculated value to another text file. That text file will be
received by AGA
to continue optimisation process (‘Data from external program (function value
for AGA)’). Default name of this file
is ‘output.txt’. This makes it possible to create a fully automated
optimisation process. In our case example, the source code looks:
#include <fstream.h>
#include <math.h>
int main(int /*argc*/, char *argv[]) {
ifstream
in(argv[1]);
ofstream
out(argv[2]);
double x1,
x2, f;
in >> x1 >> x2;
f = -5.*sin(x1)*sin(x2) -
sin(5*x1)*sin(5*x2);
out << f << endl;
return 0;
}
It calculates the value of function given by
equation (2). Source code listed above should be compiled and the executable
version should be placed into the folder together with AGA.
Input
data are received from the file specified as a first argument (argv[1]) and
result saved into file specified as a second argument (argv[2]). The AGA program
will call the external program with command-line arguments, using the syntax:
External.exe input.txt output.txt
Third
page control ‘General’ allows you to choose population size and windows you
want to see during optimisation process
-
‘Statistics
history’ – you will see statistics for every iteration or for the last one
(current) if it is unchecked
-
‘Buffer
individuals’ – to save computation time you can buffer already calculated
fitness function values. It works very well for integer and boolean variables.
-
‘Show
buffered’ – shows buffered individuals with calculated fitness function values
(if ‘Buffer individuals’ has been chosen)
-
‘Show
population’ – shows individuals of last generation. It is important for manual
delivering of fitness function. Then you have to input those values.
-
‘Population
history’ – as above (if ‘Show population’ has been chosen) but it shows all the
generations.
-
‘Show
variables’ – displays basic information about optimisation variables
-
‘(Initial)
population size’ – here you can specify the size of the population. For
variable population size method it is initial population size only (first
step).
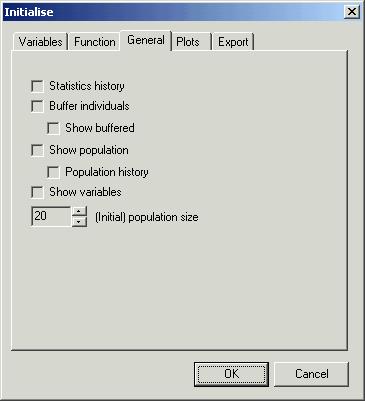
In our
case set ‘(Initial) population size’ to 30.
Fourth
page control ‘Plots’ makes possible to specify the plots you want to see during
optimisation process. One plot is predefined (for minimisation). It is plot
called ‘Convergence’. The name you choose is inconsequential. If you do not name your plot then AGA
will do it for you. To plot something you must input this in the window ‘Plot’
and push button ‘Add’. If you change your mind then remove it using button
‘Remove’.
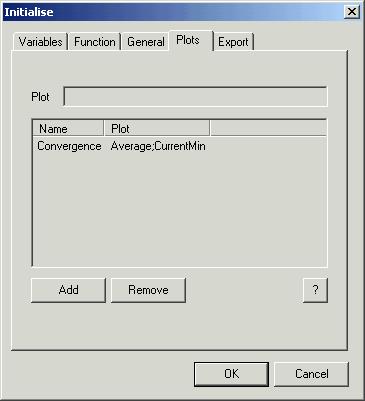
You can
plot variables or even equations. To remind yourself of the standard variable
names push button ‘?’ and you will see the following window:
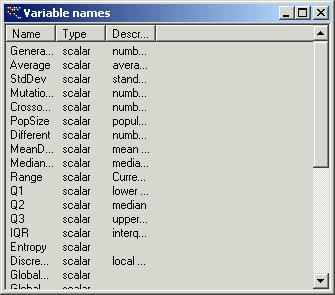
To input
your equation use variable names specified in the first column of this window.
If you want to see more than one plot in the same window the separate your
equations using ‘;’. In our case you can add two additional plots. To do it
input
Discrepancy
in the edit
window ‘Plot’, push button ‘Add’ and then change the plot’s name to
D
The second
plot is composed of two graphs that show the number of different individuals
and the function ![]() . To do it input
. To do it input
Different;2^Entropy
and
optionally change the name to
Different_vs_2^Entropy
Fifth
page control ‘Export’ allows you to export variables or equations to file. The
rules of exporting are the same as in page control ‘Plots’. There are two
differences: you have to specify a single equation (do not use ‘;’ to separate)
and you can export vector variables (instead of scalar for plotting).
To choose
the name of export file name input it to ‘Export file name’. Check box ‘Every
iteration’ allows you to export results from each iteration, or from the last
only if it is left unchecked.
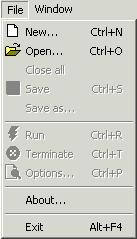
Menu
‘Window’ is composed of at least five options for windows manipulation
-
‘Cascade’
-
‘Tile
Horizontally’
-
‘Tile
Vertically’
-
‘Minimize
All’
-
‘Arrange
All’
The speed
toolbar is composed of six buttons. They are:
-
‘New’
– same as option ‘New’ form menu ‘File’
-
‘Open’
– same as option ‘Open’ from menu ‘File’
-
‘Save’
– same as option ‘Save’ from menu ‘File’
-
‘Cascade’
– same as option ‘Cascade’ from menu ‘Window’
-
‘Tile
Horizontally’ – same as option ‘Tile Horizontally’ from menu ‘Window’
-
‘Tile
Vertically’ – same as option ‘Tile Vertically’ from menu ‘Window’
-
‘Run’
– same as option ‘Run’ from menu ‘File’
-
‘Terminate’
– same as option ‘Terminate’ from menu ‘File’
-
‘Options’
– same as options ‘Options’ from menu ‘File’
![]()
If you
have decided to input function value manually then you have to supply these
values in the window ‘Population’ for each set of independent variables listed
out by the program. First click on the desired line and then click again on the
‘?’. Finally you can input a value.
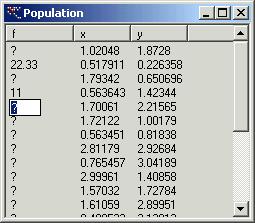
It is
also possible to modify the optimisation process options. To do this choose
either option ‘Options’ from menu ‘File’ or the relevant speed button. Dialog
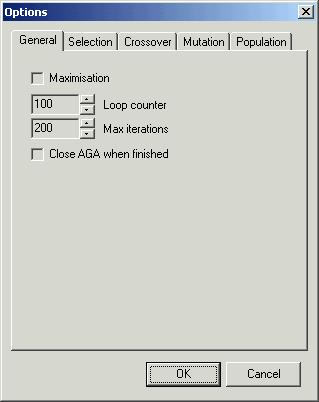
box ‘Options’ is composed of five pages. The first of them, labelled ‘General’
allows you to choose maximal number of iterations ‘Max iteration’ and change
the number of generations ‘Loop
counter’. The number of generations is important only for external and
analytical method of supplying the function value. You can decide here whether
you maximise or minimise your problem - ‘Maximisation’. AGA
may optionally be closed when the optimisation is finished. To do this choose
‘Close AGA when finished’. It is useful when you call AGA
from other program or want to fully automate your computation.
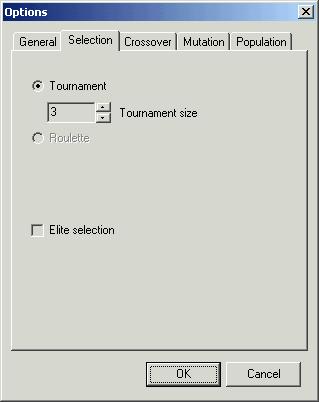
Second
page ‘Options’ allows you to choose selection method. There are two methods so
far:
-
‘Tournament’
– random tournament selection (default). You can specify the size of
tournaments ‘Tournament size’. Default tournament size equals 3. Due to its
virtues this method is recommended.
-
‘Roulette’
– roulette wheel selection. Roulette wheel method is a classical method of
individual selection. It possesses more defects than virtues.
Both
methods fits either maximisation or minimisation problems. It means that
roulette method is modified version of classical roulette.
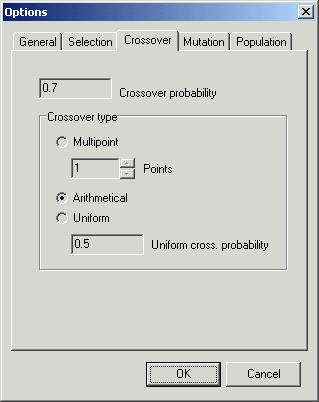
Third
page ‘Crossover’ contains options that are necessary to change crossover
method. One can also change ‘Crossover probability’. There are three crossover
methods
-
‘Multipoint’
– multipoint method. ‘Points’ indicates the number of crossover points. Default
number is 1.
-
‘Arithmetical’
– arithmetical crossover method. This method is suitable for numerical
optimisation.
-
‘Uniform’
– exchanges all the genes with ‘Uniform cross probability’
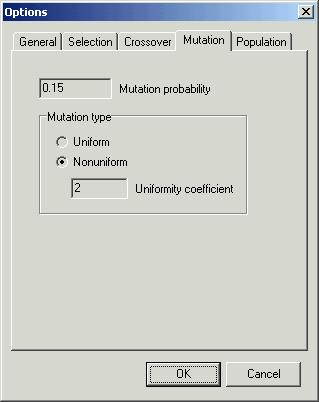
Fourth
page ‘Mutation’ allows you to specify ‘Mutation probability’ and to choose
among mutation methods
-
‘Uniform’
– this method does not depend on current generation number (stage of
optimisation)
-
‘Nonuniform’
– this method depends on current generation number. The later the generation
the smaller the influence of mutation. You can also change the ‘Uniformity
coefficient’.
Fifth
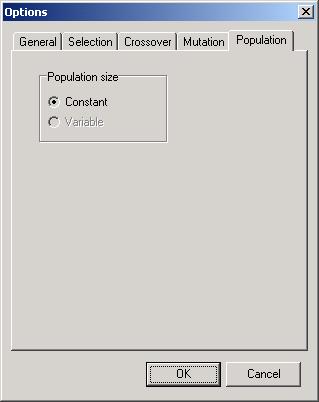
page ‘Population’ lets you to choose population size. There are two
possibilities
-
‘Constant’
– population size does not depend on generation number – is constant during
optimisation process
-
‘Variable’
– population size varies during optimisation process.
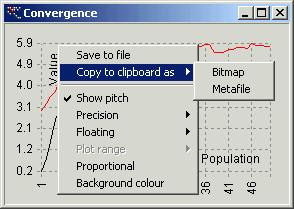
Right mouse button clicked on chart window
displays popup menu. This menu allows you to modify your chart.
-
‘Save
to file’ – save the chart as bitmap to file
-
‘Copy
to clipboard as’ – copies the chart to clipboard as
-
‘Bitmap’
-
‘Metafile’
– enhanced metafile (‘emf’ format)
-
‘Show
pitch’ – shows or hides chart pitch
-
‘Precision’
– sets number of decimal digits along specified axis
-
‘x’
-
‘y’
-
‘Floating’
– allows you to change floating number to integer on specified axis
-
‘x’
-
‘y’
-
‘Plot
range’
-
‘Proportional’
– makes the chart proportional. It is useful when x-axis and y-axis are similar
or the same
-
‘Background
colour’ – changes the background colour of your chart
I/O Console
There is a difference between capital and small
letters! When a function does not give
unique value then the main value is returned. You operate on tensors in
general. Scalars are zero valence tensors. Complex numbers are represented in
the form of
a + b*I
where ‘a’
is real part and ‘b’ imaginary part. Real parts are represented
as double precision 64 bits number. Space characters are ignored.
If input value can be
transformed into numerical value then AGA returns numerical value. For instance
2 + pi / e
gives
3.15573
if input value cannot
be transformed to numerical value then AGA will try to simplify
its part and will treat it as symbolic value. For instance
2 * pi / x
gives
6.28319/x
under condition that
you have not defined x yet.
Commands are
characterised by square brackets [] and cannot be nested.
Command list:
-
‘List[]’ – lists all declared and predefined
variables. Predefined variables are
-
pi = ![]()
-
deg = ![]()
-
![]()
-
![]()
-
Generation
– generation number
-
Average
– average fitness function
-
StdDev
– standard deviation
-
Mutations
– mutations number
-
Crossovers
– crossovers number
-
PopSize
– population size
-
Different
– number of different fitness function values
-
MeanDev
– mean deviation
-
MedianDev
– median deviation
-
Range
= CurrentMax – CurrentMin
-
Q1 –
lower quartile
-
Q2 –
median
-
Q3 –
upper quartile
-
IQR =
Q3-Q2 – interquartile range
-
Entropy
-
Discrepancy
-
GlobalMax
– maximal fitness function value from all generations
-
GlobalMin
– minimal fitness function value from all generations
-
CurrentMax
– maximal fitness function value from current generation
-
CurrentMin
– minimal fitness function value from current generation
-
CrossPoints
– number of crossover points
-
TourSize
– tournament size
-
Buffered
– number of buffered individuals
-
CrossUnifProb
– uniform crossover probability
-
CrossProb
– crossover probability
-
MutProb
– mutation probability
-
GlobalMinIndiv
– individual with minimal fitness function value from all generations
-
GlobalMaxIndiv
– individual with maximal fitness function value from all generations
-
CurrentMinIndiv
– individual with minimal fitness function value from current generation
-
CurrentMaxIndiv
– individual with maximal fitness function value from current generation
-
‘Clear[]’ – clears all declared variables
-
‘Clear[x]’ – clears
x variable
Functions
are characterised by round brackets (). Three dots in (...) mean that it is a function with variable number of arguments. All
the functions can be nested as many times as you want.
Function list:
vect(...) vect
function lets you input tensor of valence described by square number.
For instance: first valence tensor is just a vector. To obtain vector composed
of three coordinates a, b, c you must write
vect(a, b, c)
There is a shorter
and more comfortable way of inputting tensors by means of curly brackets.
Previous example now looks like this
{a, b, c}
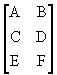
Second valence tensor
- matrix ![]() looks like this
looks like this
{{A, B, C}, {D, E, F}}
in traditional
notation
![]()
matrix ![]()
{{A, B}, {C, D}, {E, F}}
in tradition notation
sin() calculates sine function. Arguments ought
to be given in radians. To convert degrees to radians multiply them by deg. For example
sin(45 * deg)
gives ![]() or numerically
or numerically
0.707107
It is possible to
calculate sine of complex arguments or of objects different than scalars. For
instance sine of a vector
sin({I, pi, a})
gives the vector
{1.1752*I,1.22515e-16,sin(a)}
that is sine of all
components. If argument is symbolic then results are symbolic as well.
cos()
tg()
ctg()
sinh()
cosh()
tgh()
ctgh()
arcsin() calculates arcus sine of complex
argument. Result is given in radians. To convert to degrees divide it by deg. For instance for ![]() the angle equals
the angle equals ![]()
arcsin(sqrt(2) / 2) / deg
gives
45
arccos()
arctg()
arcctg()
arsinh()
arcosh()
artgh()
arctgh()
log(,) log(a, b) calculates logarithm
of b
to base a. If results of this
operation is c then a^c == b. For instance
log(2, 3)
gives
1.58496
so
2 ^ log(2, 3)
gives of course
3
One can calculates
logarithms of tensors
log(10,
{I, pi, a})
result
{0.682188*I,0.49715,log(10,a)}
that is function
log calculates logarithm of all
components. One can calculate logarithm
to different base
log({2, 3, e}, {I, pi, a})
result
{2.26618*I,1.04198,log(2.71828,a)}
The base and variable
that you calculate logarithm of have to be of the same dimension. It is because
the notation
log(10,
{I, pi})
is equivalent to
{log(10, I), log(10, pi)}
It is obvious when
you compare them
{log(10,I),log(10,pi)}
== log(10,{I,pi})
you obtain 1 as
a result (logic true)
1
ln() calculates natural logarithm. For instance
ln(2)
gives
0.693147
Notation
ln(2)
is equivalent to
log(e, 2)
compare them
ln(2) == log(e, 2)
and you shall see 1
(true)
1
exp() calculate exponential function of x ![]() . You can check it
. You can check it
e^2 == exp(2)
as a result you will
see logic 1
1
abs() calculates absolute value. For instance
abs(I)
gives
1
sqrt() calculates square root. For example
sqrt(pi)
gives
1.77245
It is the same like
pi^0.5
re() calculates real part of a complex number.
For instance
re(2+I)
gives
2
im() calculates imaginary part of a complex
number. For instance
im(2+4*I)
gives
4
arg() calculates the argument of complex number (i.e. angle in radians) where
![]() . For instance
. For instance
arg(I) / deg
gives result in
degrees
90
conj() calculates the conjugate complex number to
given argument. For instance
conj(2+I)
gives
2-I
D(,) calculates symbolic partial derivative of a
symbolic function. For instance ![]()
D(sin(x), x)
gives
cos(x)
One can calculate
composite derivative. If y equals
![]()
y = x^2
then
D(sin(y), x)
gives
cos(x^2) * 2*x
Logarithmic
derivative
D(x^x, x)
gives
x^x * (ln(x) + 1)
You can also
calculate multiple derivative. For instance ![]()
D(D(sin(x),x),x)
gives
-sin(x)
You can also
calculate derivatives of tensors.
sum(,{,,}) calculates symbolic sum. If it is
possible to give the results as a number then function sum will do it. For
instance the sum of numbers from 1 to
100 or ![]()
sum(j,{j,1,100})
equals
5050
If it is not possible
to return numerical result then the result shall be given in symbolic form. For
instance ![]() .
.
sum(x^j,{j,1,3})
gives
x + x^2 + x^3
One must take care of
sum indexes. It cannot be a variable that has already been declared. You can
calculate multiple sums by nesting one in another.
product(,{,,}) calculates symbolic product.
floor() gives the integer part of a real
number
ceil() gives the closest integer number
greater than an argument
round() gives the closest integer number of
a real number
List of operators according to priority
= assignment
|| or
&& and
!= unequal
== equal
>= greater or equal
> greater
<= less or equal
< less
- minus
+ plus
/ divide
: divide
* times
! not
~ neg (changing the sign)
^ power
{} curly brackets
() round brackets
Script writing
If first character in line is ‘;’ then the line
is treated as a comment and is ignored.
Section [VARIABLES] defines variable types and
range (if applicable). It works the same as page ‘Variables’ in dialog box
‘Initialise’. First you have to specify the name of the variable then its type
and range (if applicable). For instance
[VARIABLES]
x = real
[0; 3.1415]
z = integer
[0; 32]
b = boolean
Section [FUNCTION] defines function specifying
method: manual, analytical or external. It works almost the same like page
‘Function’ in dialog box ‘Initialise’. If you decide to supply the function by
analytical equation then you have to write
‘analytical = 1’ and then you have to specify the equation(s) in section
[FUNCTION]. If your choice is to deliver the function externally then you have
to specify also the relevant file names (see below)
[FUNCTION]
manual = 0
analytical
= 0
external =
1
input_name
= input.txt
external_name
= external.exe
output_name
= output.txt
Section [EQUATIONS] is important only if you
supply function value analytically. It works like ‘I/O Console’ All the lines
need to have variable name, character ‘=’ and finally equation. Optimised
function must be called ‘f’. You can specify as many variables as you want. For
instance
[EQUATIONS]
a
= 1
b = 5.1/4/pi^2
c = 5/pi
d = 6
e_ = 10
f_ = 1/8/pi
f = a*(x2 - b*x1^2 + c*x1 - d)^2
+ e_*(1 - f_)*cos(x1) + e_
Section [SHOW] allows you mainly to decide
which windows you want to see during optimisation process. The exception is
‘buffer_individuals’ where you decide whether to buffer individuals or not.
Option ‘arrange’ tells AGA to auto-arrange all windows.
It defines almost all the parameters form page ‘General’ in dialog box
‘Initialise’. The only exception is population size defined in section
[POPULATION].
[SHOW]
statistic_history
= 0
buffer_individuals
= 0
buffered =
0
population
= 0
population_history
= 0
variables =
0
arrange = 1
Section [OPTIONS] defines the same parameters
as page ‘General’ in dialog box ‘Options’
[OPTIONS]
maximisation
= 0
loop_counter
= 20
max_iteration
= 100
close = 0
Sections [SELECTION] defines the same
parameters as page ‘Selection’ in dialog box ‘Options’
[SELECTION]
tournament
= 1
tournament_size
= 3
roulette =
0
elite = 0
Section [CROSSOVER] defines the same parameters
as page ‘Crossover’ in dialog box ‘Options’
[CROSSOVER]
probability
= 0.7
multipoint
= 0
points
= 1
arithmetical
= 1
uniform = 0
uniform_prob
= 0.5
Section [MUTATION] defines the same parameters
as page ‘Mutation’ in dialog box ‘Options’
[MUTATION]
probability
= 0.15
uniform = 0
nonuniform
= 1
uniformity_coeff
= 2
Section [POPULATION] defines the same
parameters as page ‘Population’ in dialog box ‘Options’ except population size.
[POPULATION]
size = 30
constatnt =
1
variable =
0
Section [EXPORT_NAMES] contains some parameters
from page ‘Export’ in dialog box ‘Initialise’.
[EXPORT_NAMES]
; export_name
= export.txt
; export_every_iteration
= 1
Section [EXPORT] contains variable names and definitions (equations) you want to export. It
works like page ‘Export’ in dialog box ‘Initialise’. First you have to specify
your name then character ‘=’ and finally internal name or equation.
[EXPORT]
; gen =
Generation
; average =
Average
; standard_deviation = StdDev
; mutation_number = Mutations
; cross_num = Crossovers
; populations_size = PopSize
; diff = Different
; name1 = MeanDev
; name2 = MedianDev
; range = Range
; Q1 = Q1
; Q2 = Q2
; Q3 = Q3
; IQR = IQR
; name4 = Entropy
; D = Discrepancy
; max = GlobalMax
; min = GlobalMin
; current_max = CurrentMax
; current_min = CurrentMin
; crossover_points
= CrossPoints
; tournament_size
= TourSize
; buffered =
Buffered
; uniform_cross_probability
= CrossUnifProb
; cross_probability
= CrossProb
; mutation_probability
= MutProb
;
; a =
GlobalMinIndiv
; b =
GlobalMaxIndiv
; c =
CurrentMinIndiv
; d =
CurrentMaxIndiv
;
; size =
2^Entropy
Section [PLOTS] defines the same parameters as
page ‘Plots’ in dialog box ‘Options’. First you must specify name of the window
then character ‘=’ and finally variable name or equation. If you want to see
more graphs in one window then separate your equations by means ‘;’.
[PLOTS]
Convergence
= Average; CurrentMin
; name1 =
2^Entropy
; iqr = Q3-Q1
quartiles =
Q1; Q2; Q3
; name4 =
PopSize; Different; 2^Entropy
Download AGA for
non-commercial use only.